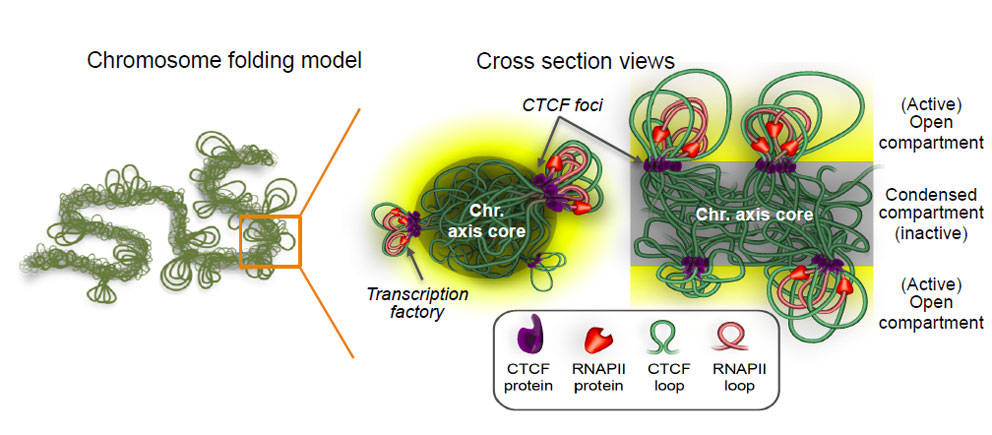
Protein factors are responsible for organizing chromosomes inside the nucleus in three dimensions (3D), forming a shape like a gift bow, with proteins aggregating as the central “knot” holding the ribbon-like loops of DNA when genes are organized for proper expression, or a tangled mess in the presence of certain mutations.
These findings, published in Cell, result from research by an international team of collaborators, led by The Jackson Laboratory, exploring the 3D genome architecture in the nucleus and its relationship to gene regulation and diseases.
Using advanced 3D genome mapping technologies, 3D genome simulation and super-resolution microscopy, the researchers identified the topological framework mediated by chromatin proteins (CTCF and cohesin), in which genes are organized and transcribed at the chromosomal level. They found that the protein-bound knots were located along the chromosome axis, with the DNA loops extending out from the transcriptionally inactive chromosome cores. This is the first observation of the barrier between genes being actively read and those that are not.
They also determined that the CTCF/cohesin-mediated genome organization is not random. For instance, “housekeeping genes” (those essential genes commonly expressed in all cell types) were often found near the protein-bound knots, whereas cell-specific genes were more commonly positioned in the extended DNA loops. Mechanistically, depending on the specific needs of a cell, transcriptional proteins selectively draw certain genes on those loops close to the protein knots for cell-specific expression.
Moreover, they uncovered a potential mechanistic link between genetic mutations and their associated disease. A single nucleotide mutation associated with asthma and autoimmune disease in people of European ancestry disrupted CTCF-DNA binding, the topology of the DNA looping and subsequent transcription of the surrounding genes, resulting in a messy tangle of DNA instead of a neat bow. Thus, protein-DNA binding data may prove useful for determining which specific nucleotide changes in the human genome, through altering genome topology, are responsible for driving disease.
This research was supported in part by National Institutes of Health grants NCI R01 CA186714, NHGRI R25HG007631 and NIDDK U54DK107967, and the JAX Director’s Innovation Fund.