New research about centromere DNA sequence and protein interactions in four diverse mouse strains have implications that may change the way we view the centromere, cell division and genomic instability-caused conditions.
A (centromere) butterfly effect
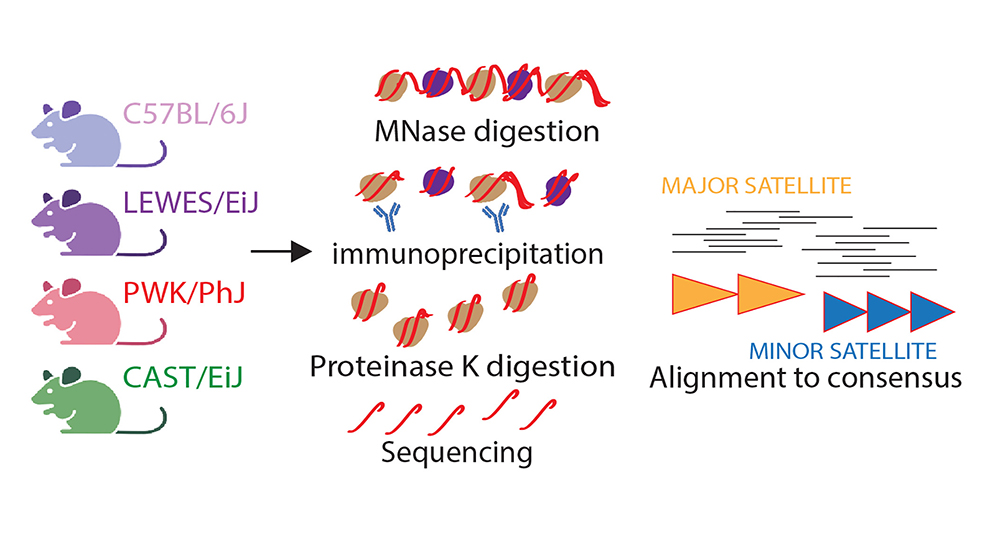
Responsible for giving chromosomes their distinct shape, centromeres are also essential genetic stabilizers. Loss of chromosome integrity can lead to sudden cell death and mistakes in cell division. These tiny slipups have the potential to become widespread genomic errors that can drive cancer formation or cause infertility. As part of The Jackson Laboratory, Associate Professor Beth Dumont, Ph.D., explores the field of evolutionary genetics. In recent research, Dumont documented the centromere DNA sequence and protein interactions in four diverse mouse strains. Recently published in Cell Reports, her findings have implications that may change the way we view the centromere, cell division and genomic instability-caused conditions.
Divide and conquer
We rely on cell division to grow, maintain and repair our cells and tissues. One of the most crucial aspects of this process is chromosome segregation (where jumbled, threadlike DNA condenses to form chromosome copies for the new cells being formed). Dumont recognized a key player in this segregation process is a protein complex called a kinetochore, which attaches to centromere DNA to separate chromosome copies into their respective daughter cells. She analyzed a kinetochore protein called CENP-A and found that the locations where it positioned and attached itself to the centromere were highly variable. The distinct sequence-protein interactions identified by the Dumont lab allow for further pattern recognition analysis of when, where and how chromosome segregation errors occur. By identifying a predictable map of these interactions, researchers like Dumont can determine what it takes to complete successful chromosome segregation, as well as what introduces errors that result in genomic instability.