
Introduction
One of the major challenges oncologists are often faced with when managing precision cancer treatment is acquired drug resistance. Resistance due to emerging gene variants can lead to progressive disease, likely requiring treatment with second- or third-line therapies. Identifying drugs to bypass known resistance mechanisms or drug combinations to target the initial driver and co-occurring resistance variants remains a hurdle, limiting the long-term success of precision medicine. Preclinical approaches to new drug development or even new drug combinations may include testing cancer cell lines with a panel of known resistance variants. However, with the vast amount of literature out there, the task of finding variants that potentially contribute to drug resistance is not only time consuming but at times, overwhelming. The JAX Clinical Knowledgebase (CKB) is an invaluable resource that can be used to aid in identifying a list of potential resistance variants for preclinical testing (Patterson et al. 2016).
Example of a use case- targeting the MAPK/ERK pathway
The MAPK/ERK pathway is responsible for regulation of cell growth and proliferation and situated in the middle of the pathway are the RAF genes, ARAF, BRAF, and CRAF, which activate the pathway through phosphorylation of MAP2K1/2 and ERK. While mutations in the various pathway components can lead to uncontrollable cell growth (Zhang and Liu, 2002), the gene most commonly mutated is BRAF. Fortunately, since 2011, several RAF/BRAF-targeting therapies have been FDA approved for use in multiple tumor types including melanoma, non-small cell lung cancer, and thyroid cancer. These drugs, such as Vemurafenib, Dabrafenib, and Encorafenib, act to inhibit BRAF and subsequent downstream signaling, resulting in antitumor activity (Patel et al. 2020).
Reactivation of the pathway downstream of BRAF via secondary mutations can result in drug resistance, leading to tumor growth and disease progression (Olbryt, 2022). Therefore, when developing a new drug or considering the use of a new combination of drugs, it is important to consider the potential for resistance variants and whether they can bypass treatment. Preclinical testing of cell line panels expressing resistance variants can provide a preview of what might translate to the clinic.
One approach to developing such a panel is to find case studies that report variants detected in patients following progression on RAF/BRAF targeted therapies. The challenge with this method is that individual patients may present with a molecular profile consisting of several variants in different genes. Tumors are often heterogenous and you may find clonal effects during sequencing. Trying to parse out the true resistance mechanism can be difficult and is typically untested in the clinical arena. In this case, preclinical data can be of significant value – by using cell lines with a tested and clearly defined molecular profile, you can identify variants that drive resistance rather than observing variants after resistance has occurred.
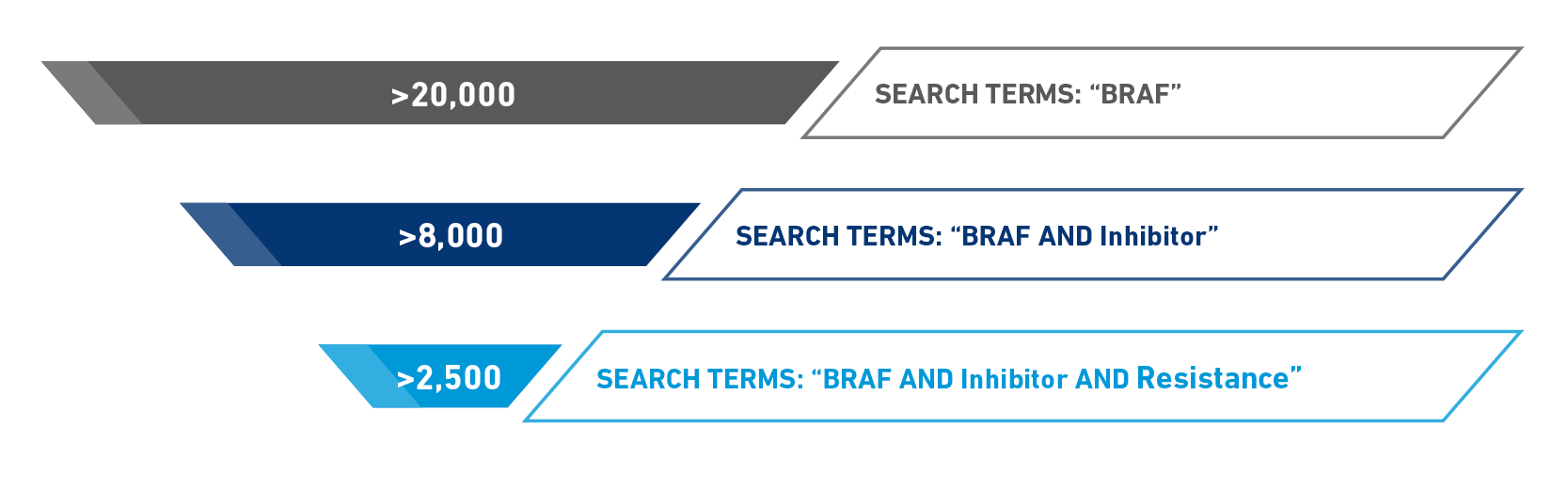
Manual searching through either PubMed or Google Scholar for resistance variants may lead to more results than you can reasonably sort through (Figure 1), and even if you can narrow the results down further, you will need to read through each paper to identify the resistance variants and methodology that was used. This type of searching also requires cross referencing to make sure you do not miss anything while simultaneously finding many articles referencing the same variants. This can be onerous, time consuming, and inefficient.
Figure 1: PubMed results for different combinations of search terms for BRAF inhibitor resistance variants
However, with CKB, you not only save valuable time but also have the ability to effectively narrow down or expand your search depending on your needs. CKB is a structured database that provides summary level data for thousands of variants, which is sortable and filterable – using controlled vocabulary to help you efficiently develop a list of potential resistance variants. Additionally, all the data come from peer-reviewed published literature, and links out to those publications, so you can always go to the original paper for additional information.
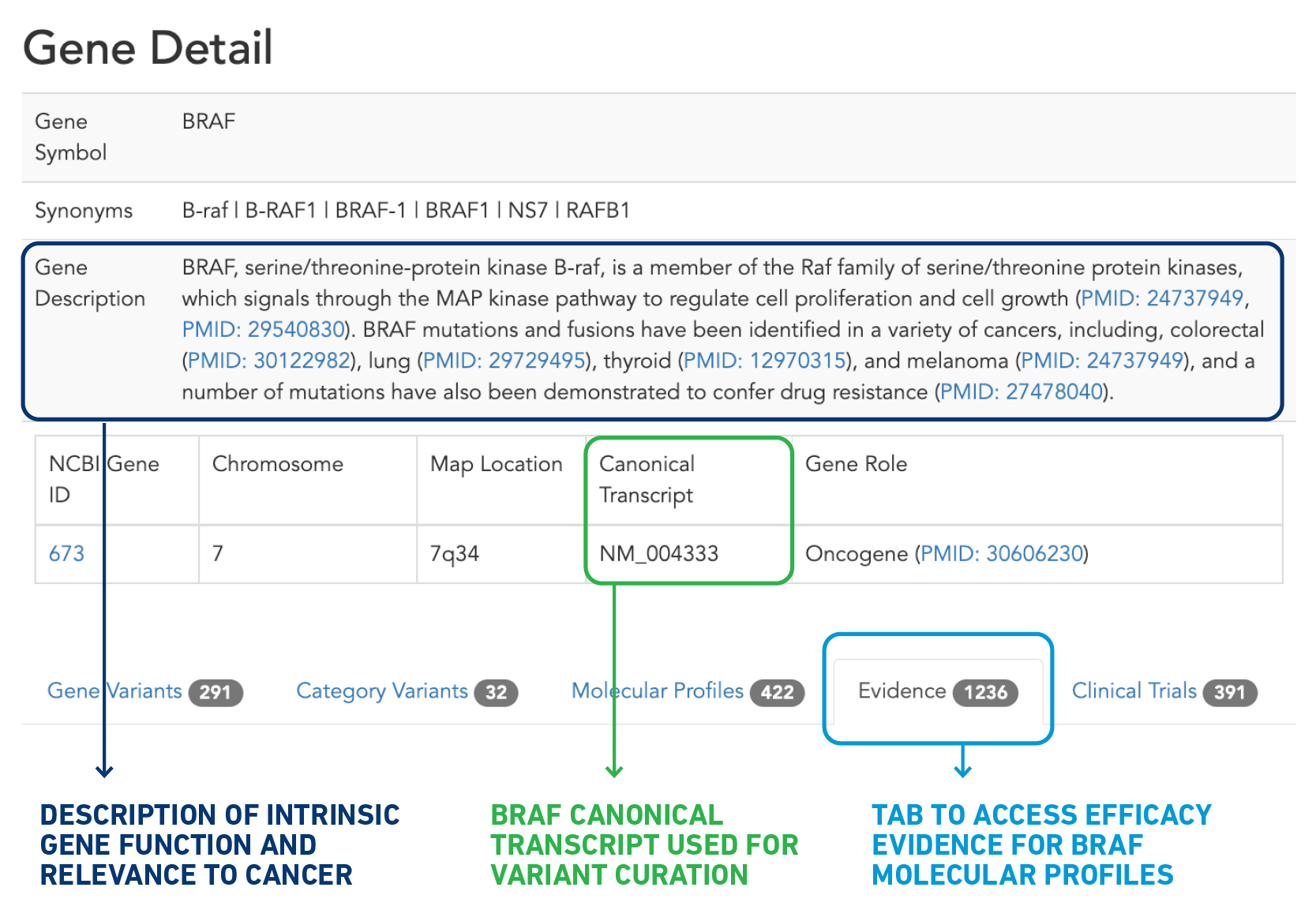
There are several ways to search for resistance variants, but let’s walk through one approach. After navigating to the BRAF gene detail page via “Explore by Gene” on the CKB home page (ckb.jax.org), you’ll notice several areas of interest on this page (Figure 2). The top portion provides you with basic information about the gene, a description of the intrinsic function of the gene and its relationship to cancer, and the canonical transcript used in CKB. Farther down the page is a table with several tabs, the last of which is “Gene Level Evidence”. This tab provides all the efficacy evidence, which connects a response to a therapy and molecular profile (consisting of one or more variants) that has been curated with at least one BRAF variant.
Figure 2: Anatomy of a Gene Detail page.
When you initially click on the tab, the table will be sorted by approval status with the highest level of evidence at the top – FDA approvals and Guidelines. While you can scroll down to find the preclinical-level efficacy evidence, the easiest way to find it is to use the filter box at the top of the approval status column. Additional columns include molecular profile, indication/tumor type, response type, therapy name, and evidence type. You can sort and/or filter on multiple columns at the same time, allowing you to find the specific data you are interested in. As you filter down, you will see the number of returned entries changes, giving you an idea of the amount of data available to sort through.
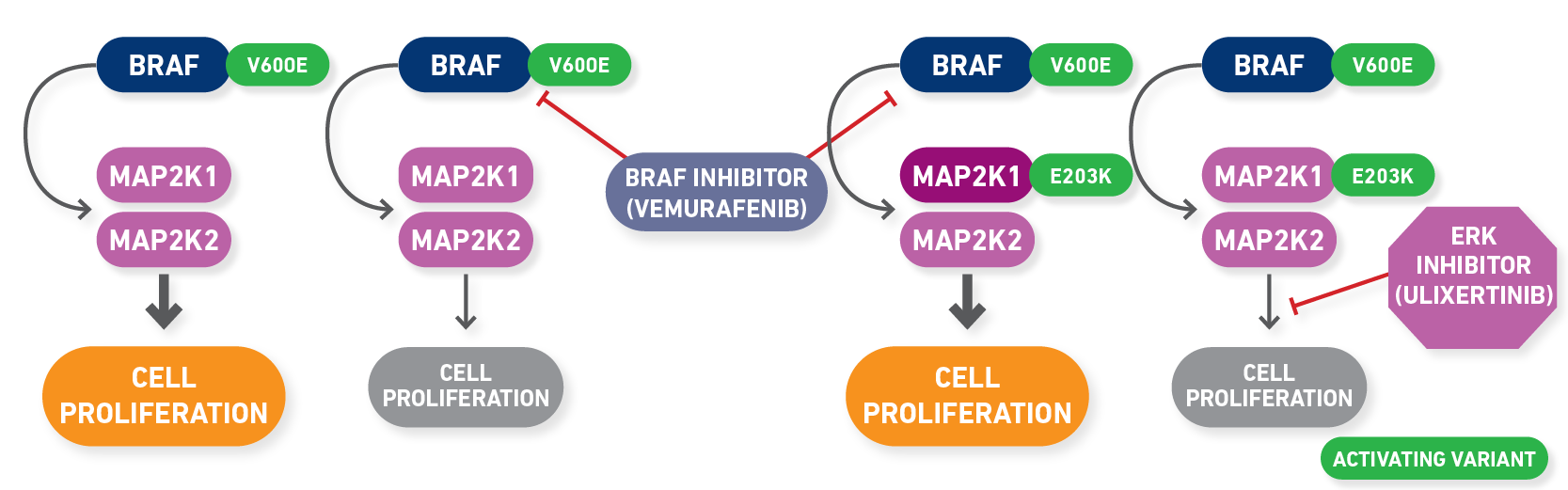
As you scroll through the rows of evidence after filtering on preclinical and resistant, you may notice another gene that is frequently mutated with BRAF mutations, MAP2K1. MAP2K1 encodes MEK1, which lies downstream of BRAF in the MAPK/ERK pathway, and therefore, variants in MAP2K1 can result in secondary resistance to BRAF inhibitors by reactivating the pathway downstream of BRAF inhibition (Olbryt, 2022). Searching specifically for MAP2K1 in the “Molecular Profile” column will filter down to evidence associated with these resistance variants, which may be the start of your panel of preclinical resistance variants (Figure 3).
Figure 3: Searching specifically for MAP2K1 in the “Molecular Profile” column will filter down to evidence associated with these resistance variants.
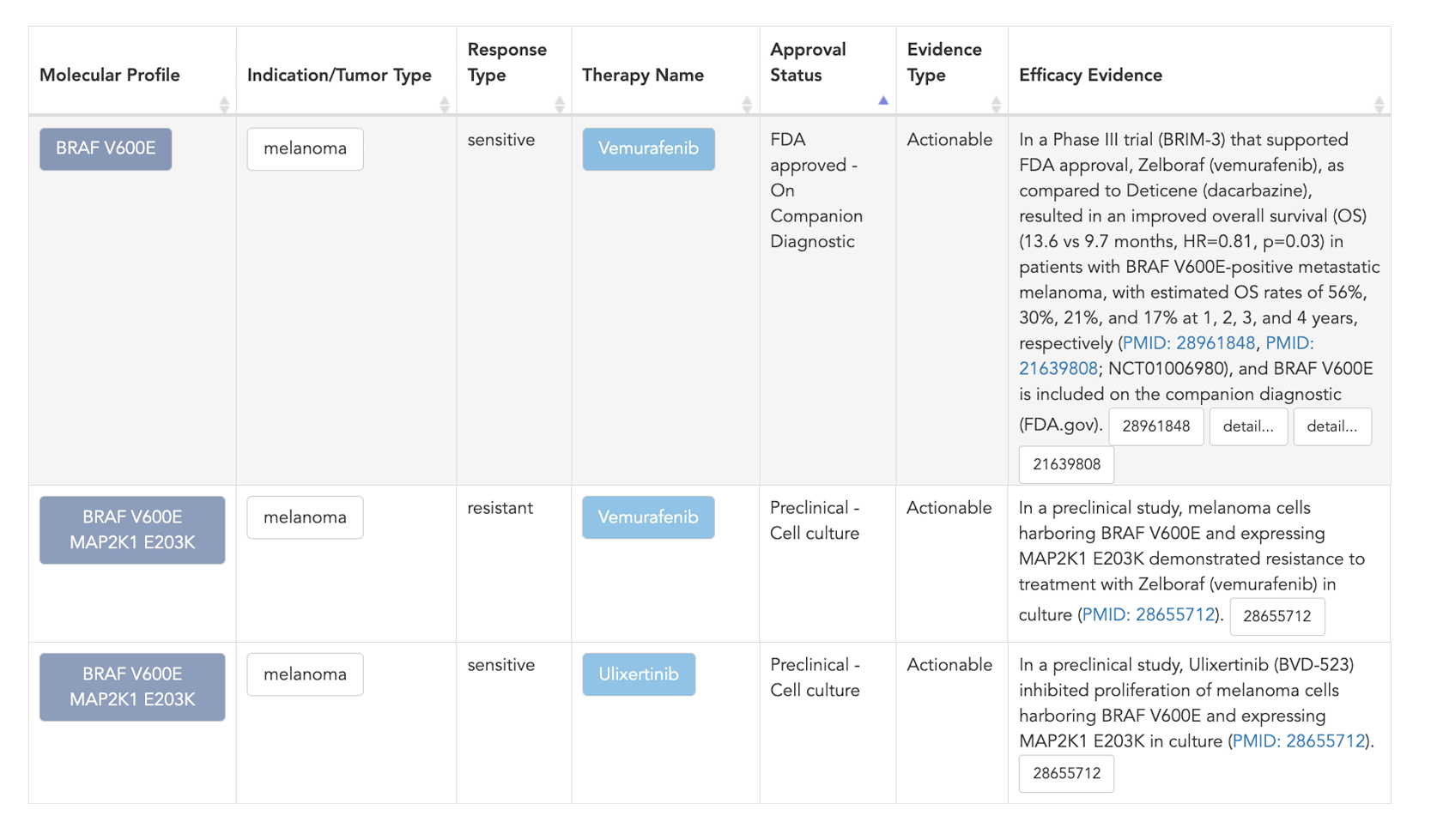
After filtering the data, you can review the efficacy evidence line for each molecular profile, therapy, and indication. From here you can go in many directions to find additional information. You can click on a molecular profile to find all evidence that is associated with that profile. You can explore a specific therapy or indication. If you want information about how the models were derived, you can click on the reference, which will direct you to the article in PubMed, allowing you to review the primary data and methods.
Conclusion
In the context of cancer research and drug development, access to a well-structured knowledgebase such as CKB saves time and provides an accurate and comprehensive view of the preclinical landscape. While this specific use case is one such example of how CKB can be of benefit, please feel free to reach out to us at ckbsupport@jax.org if you have any additional questions or want to discuss other ways CKB may be of value to your preclinical needs.
References
Olbryt M. Potential Biomarkers of Skin Melanoma Resistance to Targeted Therapy-Present State and Perspectives. Cancers (Basel). 2022 May 6;14(9):2315. doi: 10.3390/cancers14092315. PMID: 35565444; PMCID: PMC9102921.
Patel H, Yacoub N, Mishra R, White A, Long Y, Alanazi S, Garrett JT. Current Advances in the Treatment of BRAF-Mutant Melanoma. Cancers (Basel). 2020 Feb 19;12(2):482. doi: 10.3390/cancers12020482. PMID: 32092958; PMCID: PMC7072236.
Patterson SE, Liu R, Statz CM, Durkin D, Lakshminarayana A, Mockus SM. The clinical trial landscape in oncology and connectivity of somatic mutational profiles to targeted therapies. Hum Genomics. 2016 Jan 16;10:4. doi: 10.1186/s40246-016-0061-7. PMID: 26772741; PMCID: PMC4715272.
Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002 Mar;12(1):9-18. doi: 10.1038/sj.cr.7290105. PMID: 11942415.