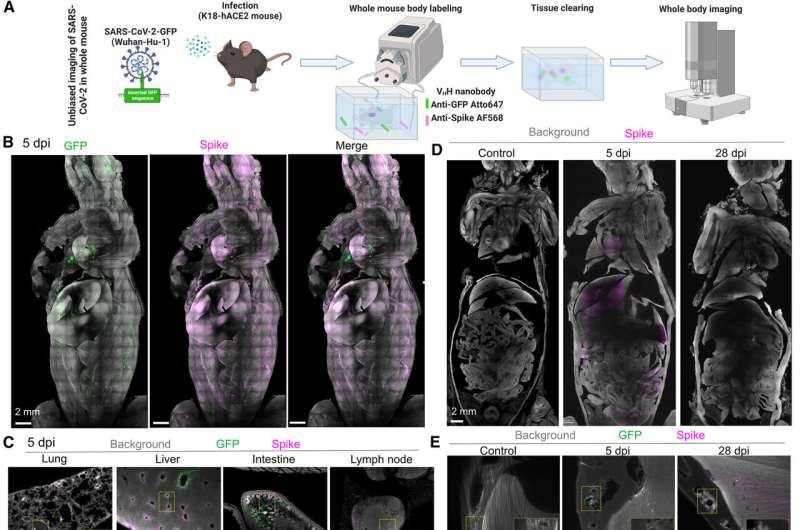
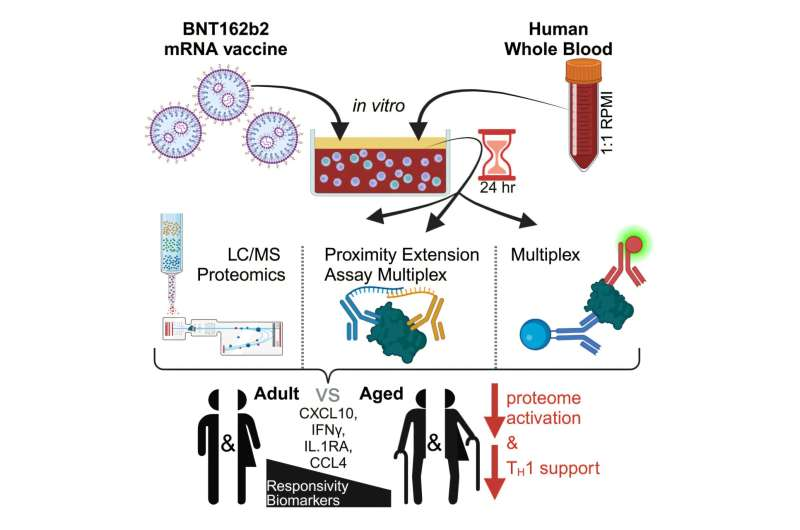
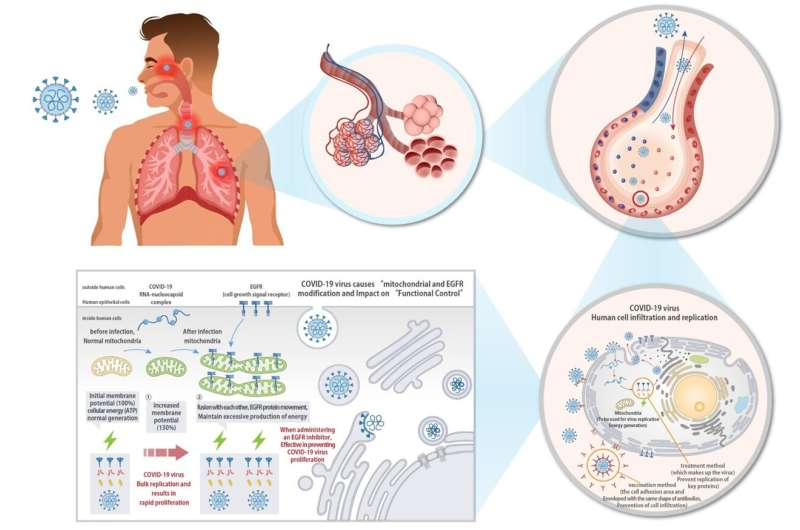
Coronavirus Research using the Laboratory Mouse
The laboratory mouse is an important preclinical model for studying coronavirus biology and treatment approaches. The information on this site summarizes information about mouse strains and genes relevant to the study of coronavirus infection and pathology. This information has been expertly curated from pre-prints and peer-reviewed publications dating back to the 1980s by biocuration scientists from the Mouse Genome Informatics (MGI) knowledgebase hosted at The Jackson Laboratory.
See all curated publications that report on mouse models and genes related to coronaviruses in MGI.
Mouse Strains Used in Coronavirus Research
Mouse strains used in coronavirus research with associated mouse phenotype categories and human disease terms and references citing the use of the mouse strains in coronavirus research. Under each reference is a list of keywords for the research areas and the viruses mentioned in the publication. A virus name appended with ‘MA’ indicates that the virus is a mouse adapted strain.
Mouse strain names are linked to strain detail pages at MGI. Human disease terms are linked to the Disease ontology resource at MGI. References are linked to the reference detail pages at MGI.
Loading...
Alleles for Laboratory Mouse Strains Used in Coronavirus Research
Alleles for laboratory mouse strains used in coronavirus research along with mouse phenotype categories and human disease terms and relevant references. Under each reference is a list of keywords for the research areas and the viruses mentioned in the publication. A virus name appended with ‘MA’ indicates that the virus is a mouse adapted strain.
Mouse alleles names are linked to allele detail pages at MGI. Human disease terms are linked to the Disease Ontology Browser at MGI. References are linked to the reference detail pages at MGI.
Loading...
Mouse Genes Associated with Coronavirus Infection and Pathology
Curated set of mouse genes associated with coronavirus infectious disease and pathology with links to the relevant reference(s). Under each reference is a list of keywords for the research areas and the viruses mentioned in the publication. A virus name appended with ‘MA’ indicates that the virus is a mouse adapted strain.
Genes are linked to the gene detail pages at the Mouse Genome Informatics (MGI) database and to the mouse gene page at the Alliance of Genome Resources. References are linked to the reference detail pages at MGI.
Loading...
Human Genes Associated with Coronavirus Infection and Pathology
This table lists human genes implicated in coronavirus infectious disease and pathology with links to the supporting publications. Instances where genes are not implicated are also included in the table. Data for this table are imported from the Alliance of Genome Resources. Only gene associations with Coronavirus infectious disease (DOID:0080599) or any of its descendants are shown. Links to mouse orthologs of the human genes are provided.
Gene symbols and disease terms link to the Alliance of Genome Resources web site. References link to the PubMed web site. Mouse ortholog symbols link to MGI gene detail pages.
Loading...
About SARS-CoV-2
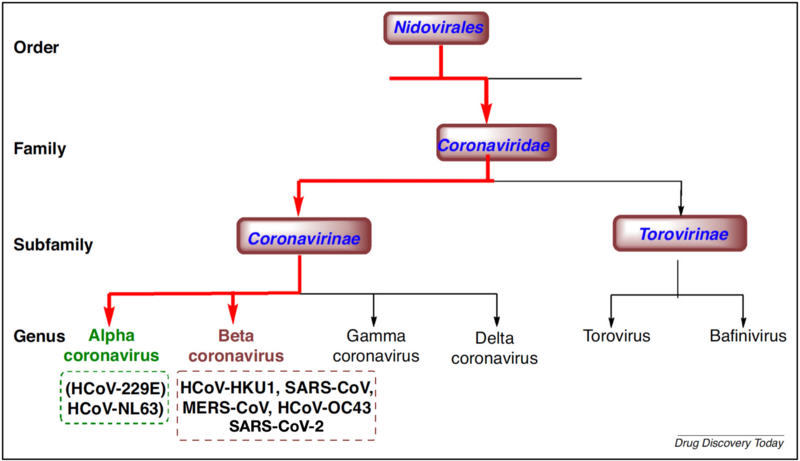
SARS-CoV-2 is positive single strand RNA virus and is a member of the Coronaviridae family of viruses which are known to target the respiratory systems of mammals.
There are four genera within Coronaviridae: alpha, beta, delta, and gamma. Only alpha and beta genera are transmissible to humans.
Although the genome of SARS-CoV-2 is similar to the zoonotic coronaviruses SARs-CoV and MERS-CoV that caused the SARs outbreak of 2003 and the MERS outbreak of 2012, the emerging evidence suggests that SARS-CoV-2 did not descend from SARS-CoV. SARS-CoV-2 causes a different range of diseases in humans and has a different transmission efficiency.
SARS-CoV-2 was likely transmitted to humans from an animal reservoir. The exact nature of this transmission is still under investigation.
Other Research Resources