Settings
Save and close
The Jackson Laboratory maintains and distributes chromosome-aberration stocks that provide mouse models for Down syndrome, as well as the study of chromosomal aneuploidy. With over 100 strains for the study of Down Syndrome and other chromosomal anomalies, the JAX® Mice Repository houses the largest collection in the United States. Each mouse autosome is present in at least two Robertsonian chromosomes,so that these stocks can be used to generate specific aneuploidies for each of the 19 mouse autosomes.
The cytogenetics and Down syndrome resource is funded by an NICHD/NIH contract; users are required to fill out a request form for mice. Please complete and submit the request form to NIH. Your order must be placed with our Customer Service Department by phone at 1-800-422-MICE or (207) 288-5845, or by fax to 207-288-6150.
This resource is supported by NICHD contract #275201000006C-3-0-1. Please reference this funding from the National Institute of Child Health and Human Development/NIH in citations acknowledging use of the resource.

A team led by David Serreze presents a mouse model for studying severe immune checkpoint inhibition-based damage to heart and skeletal muscle, providing an important resource for maximizing the benefits and minimizing the side effects of vital immunotherapies.

JAX scientists are working to make it more feasible for researchers to incorporate diverse background genetics to better model human disease in mice.

A new protocol sets the stage for researchers to directly compare mouse and human cells, and readily incorporates genetic diversity into mouse-based research to more closely approximate human health conditions.

Two world-leading organizations are partnering to further expand AI capabilities in biomedical science and catalyze medical progress.
View more
JAX researchers have created a panel of genetically diverse mice that accurately model the highly variable human response to SARS-CoV-2 infection. This will allow scientists to model patient variation in COVID-19 outcome.
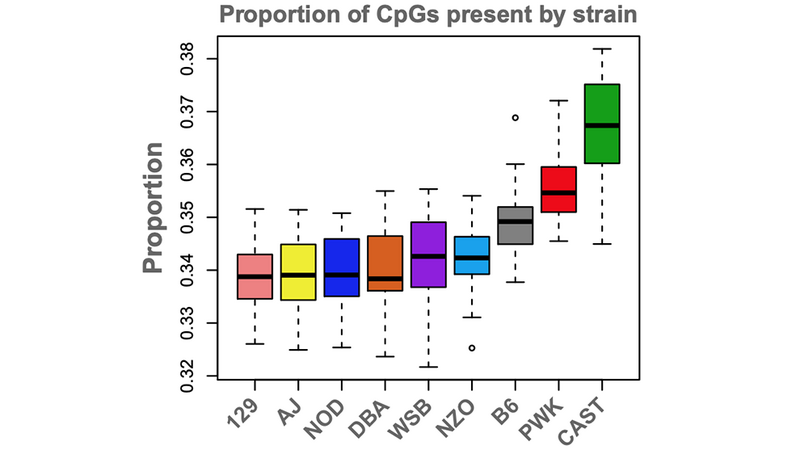
New research finds that the epigenetic landscape is highly variable between the strains of Diversity Outbred mice and is associated with variation in gene expression.

James Godwin has received a a five-year, $2.4M R01 grant to investigate regeneration in model organisms, and his work with mice so far has shown that multiple immune-cell types inhibit tissue regeneration.